two equivalent phylogenetic trees drawing

To better understand what a phylogeny represents, start by imagining one generation of butterflies of a particular species living the same area and producing offspring. If you focus on four individual butterflies in both the parental and offspring generations, the resulting pedigree may appear like the one in Figure 1B.
Now, expand your image to encompass all the butterflies of this species in a particular meadow over several generations. A pedigree for this population might look something like the one in Figure 1C. Note that each individual in the figure has two parents, but each gives rise to a variable number of offspring in the next generation.
Next, imagine taking your pedigree and getting rid of the organisms, thus keeping only the descent relationships, which are the glue that holds the population together (Figure 1D inset box). Then zoom out even farther to include many more individuals (say, from multiple meadows in the same region) and more generations. For example, the whole of Figure 1D is derived from a similar diagram as the inset box, but it now includes more individuals and many generations. As you can see, if one were to try to represent a typical population of several thousand individuals that persists for hundreds or thousands of generations, all one would see would be a fuzzy line.
Individual populations may be fairly isolated for some period of time. However, on an evolutionary timescale, migration will occur among the discrete populations that make up a typical species. This gene flow between populations has the effect of "braiding" the population lineages into a single species lineage, which might be thought of as resembling Figure 1E.
Moreover, during evolution, lineages often split. This occurs when populations or groups of populations become genetically isolated from one another. Lineages most commonly split because of the migration of a few individuals to a new, isolated region (e.g., an island). This is sometimes called a founder event. Alternatively, a formerly contiguous range can be broken up by geological or climatic events (e.g., the creation of mountains, rivers, or patches of inhospitable terrain). This phenomenon is called vicariance. No matter whether populations split due to founder events or vicariance, if the isolated populations remain separate, they will start evolving differences from one another (Figure 1F). After all, a mutation that arises in one population will have no way to get to the other population. Thus, even a mutation that would be selectively favored in both populations will become fixed in only one of the groups.
As a consequence of this genetic isolation, the lineages will evolve separately, becoming more and more different over time. If they remain apart for long periods, enough physiological and behavioral differences may evolve to result in reproductive isolation, such that it will be impossible for individuals from the two lineages to reproduce even in the case that they do come back into contact. Because of this, it is a useful simplification to assume that once lineages diverge, the two sets of descendants will remain distinct.
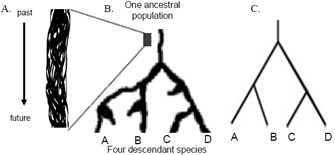
Figure 2B shows what we might see if we followed the fate of a single ancestral lineage (Figure 2A) long enough that it gave rise to four descendant lineages (species). This example includes three lineages that were established but became extinct before the end of the observation period. This diagram is an example of a simple phylogenetic tree.
In most cases, researchers draw phylogenetic trees in such a way as to record only those events that are relevant to a set of living taxa. Most commonly, these taxa are species. For example, Figure 2C shows the basic tree we could draw to represent the history of the four "tip" species, A through D. This tree shows that species A and B share a more recent common ancestor with each other than with either species C or species D. Likewise, species C and D share a more recent common ancestor with each other than with either species A or species B. This example illustrates the fact that a phylogeny is, at its most basic level, a history of descent from common ancestry.
Phylogenetic trees are fractal in the sense that the same pattern is found whether we consider recently diverged lineages or deep splits in the tree of life. Indeed, the most basic postulate of evolutionary theory is that the same general phenomenon of descent from common ancestry applies to both the most diverse branches of the tree of life and the most similar. As a result, the tree structure is extremely helpful in tracking biological diversity at all levels.
Source: http://www.nature.com/scitable/topicpage/reading-a-phylogenetic-tree-the-meaning-of-41956
0 Response to "two equivalent phylogenetic trees drawing"
Post a Comment